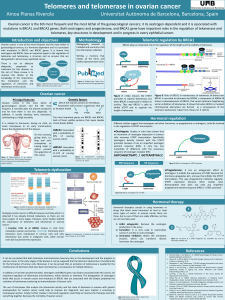
Clinical characteristics and outcomes of patients with BRCA1 or RAD51C
methylated versus mutated ovarian carcinoma
Sarah S. Bernards
a
,Kathryn P. Pennington
a
,Maria I. Harrell
a
,Kathy J. Agnew
a
,Rochelle L. Garcia
b
,
Barbara M. Norquist
a
,Elizabeth M. Swisher
a,
⁎
a
Division of Gynecologic Oncology, Department of Obstetrics and Gynecology, University of Washington, 1959 Northeast Pacific Street, Seattle, WA 98195, USA
b
Department of Pathology, University of Washington, 1959 Northeast Pacific Street, Seattle, WA 98195, USA
HIGHLIGHTS
•Methylation and mutation of RAD51C or BRCA1 were mutually exclusive.
•Methylation of BRCA1 was associated with younger age at diagnosis.
•Germline BRCA1 mutations and BRCA1 methylation were associated with HGS histology.
abstractarticle info
Article history:
Received 22 July 2017
Received in revised form 28 November 2017
Accepted 4 December 2017
Available online xxxx
Objective. In ovarian carcinoma, mutations in homologous recombination DNA repair (HRR) genes, including
BRCA1 and RAD51C, are associated with increased survival and specific clinical features. Promoter hypermethy-
lation is another mechanism of reducing gene expression. We assessed whether BRCA1 and RAD51C promoter
hypermethylation is associated with similar survival and clinical characteristics.
Methods. Promoter methylation of BRCA1 and RAD51C was evaluated using methylation-sensitive PCR in 332
primary ovarian carcinomas. Damaging germline and somatic mutations in 16 HRR genes were identified using
BROCA sequencing.
Results. BRCA1 methylation was detected in 22 carcinomas (6.6%) and RAD51C methylation in 9 carcinomas
(2.7%). These small numbers limited the power to detect differences in survival and platinum sensitivity. Mutations
in one or more HRR genes were found in 95 carcinomas (29%). Methylation of BRCA1 or RAD51C was mutually
exclusive with mutations in these genes (P= 0.001). Patients whose carcinomas had BRCA1 methylation
(57.7 years ± 2.5) or BRCA1 mutations (54.1 years ± 1.4) were younger than those without (63.3 years ± 0.8;
P=0.029,Pb0.0001). BRCA1 methylation and germline BRCA1 mutation were associated with high grade serous
(HGS) histology (P=0.045,P=0.001).BRCA1 mutations were associated with increased sensitivity to platinum
chemotherapy while BRCA1 methylation was not (P= 0.034, P= 0.803). Unlike HRR mutations, methylation
was not associated with improved overall survival compared to cases without methylation or mutation.
Conclusions. Patients with BRCA1-methylated carcinomas share clinical characteristics with patients with
BRCA1-mutated carcinomas including younger age and predominantly HGS histology. However, unlike mutation,
RAD51C and BRCA1 methylation were not associated with improved survival or greater sensitivity to platinum
chemotherapy.
© 2017 Published by Elsevier Inc.
Keywords:
Ovarian carcinoma
Methylation
Homologous recombination
Mutation
1. Introduction
Ovarian, fallopian tube and primary peritoneal (collectively called
ovarian) carcinomas commonly have deleterious mutations in the ho-
mologous recombination DNA repair pathway (HRR) genes BRCA1 and
BRCA2 (BRCA1/2), with germline mutations in approximately 15% of pa-
tients and somatic mutations in another 6% of carcinomas [1–4].
Germline mutations in these genes are the most common cause of
inherited ovarian carcinoma and are associated with increased
Gynecologic Oncology xxx (2017) xxx–xxx
⁎Corresponding author at: University of Washington Medical Center, Department of
Obstetrics and Gynecology, Box 356460, Seattle, WA 98195-6460, USA.
E-mail addresses: sarahb[email protected] (S.S. Bernards), katypenn@uw.edu
(K.P. Pennington), maribel@uw.edu (M.I. Harrell), kagnew@uw.edu (K.J. Agnew),
rochelle@uw.edu (R.L. Garcia), bnorquis@uw.edu (B.M. Norquist), swishere@uw.edu
(E.M. Swisher).
YGYNO-976984; No. of pages: 5; 4C:
https://doi.org/10.1016/j.ygyno.2017.12.004
0090-8258/© 2017 Published by Elsevier Inc.
Contents lists available at ScienceDirect
Gynecologic Oncology
journal homepage: www.elsevier.com/locate/ygyno
Please cite this article as: S.S. Bernards, et al., Clinical characteristics and outcomes of patients with BRCA1 or RAD51C methylated versus mutated
ovarian carcinoma, Gynecol Oncol (2017), https://doi.org/10.1016/j.ygyno.2017.12.004

sensitivity to platinum chemotherapy, better five-year survival, high
grade serous (HGS) histology, and somatic TP53 mutations [5–8]. Our
group and others have recently shown that somatic mutations in
BRCA1/2 and some non-BRCA1/2 HRR genes including RAD51C are also
associated with platinum sensitivity, improved survival, and sensitivity
to poly (ADP-ribose) polymerase (PARP) inhibitors [2,9–11].
Promoter hypermethylation (methylation) is another biologic
mechanism of reducing gene expression and occurs frequently in cancer
[12].BRCA1 promoter methylation is common in ovarian carcinoma
with rates from 8 to 15% and is associated with both reduced RNA and
protein expression [12–19]. In HGS ovarian carcinomas, The Cancer Ge-
nome Atlas (TCGA) identified BRCA1 and RAD51C as the only HRR genes
in which promoter methylation correlated with reduced RNA expres-
sion [1].RAD51C methylation has been described in 1–3% of ovarian car-
cinomas [1,15].
Given the improved outcomes seen with BRCA1 mutation and the
functional impact of BRCA1 promoter methylation (decreased RNA
and protein expression), we hypothesized that BRCA1 and RAD51C
methylation would also be associated with improved outcomes
and similar clinicopathological characteristics. However, there
are conflicting reports of the prognostic value of BRCA1 methylation
in regards to sensitivity to platinum chemotherapy and survival
[1,14,15,20,21]. No previous study except TCGA, which was limited
only to ovarian carcinomas of HGS histology, has considered both
methylation and mutation in many HRR genes in comparing out-
comes [1,14,15,20,21]. We sought to evaluate BRCA1 methylation
and RAD51C methylation in ovarian carcinomas of varying histolo-
gies and compare outcomes and characteristics of methylated versus
mutated cases.
2. Methods
Patients with ovarian, fallopian tube, or peritoneal carcinoma pro-
vided IRB-approved informed consent to enroll in the University of
Washington gynecologic oncology tissue bank at the time of their pri-
mary debulking surgery. Patients diagnosed with carcinoma at the
time of planned prophylactic bilateral salpingo-oophorectomy were ex-
cluded. Germline and neoplastic DNA was sequenced using BROCA, a
targeted capture and massively parallel sequencing platform previously
described [2,22,23].
Carcinomas with a damaging germline or somatic mutation in ATM,
ATR,BARD1,BLM,BRCA1,BRCA2,BRIP1,CDK12,CHEK2,MRE11A,NBN,
PALB2,RAD51C,RAD51D,RBBP8,SLX4,orXRCC2 were classed as having
HRR deficiency.
Methylation of RAD51C and BRCA1 in neoplastic DNA was assessed
by bisulfite conversion using the Zymo Research EZ DNA Methylation-
Direct kit followed by methylation-specific PCR as previously described
[9,17]. Methylation was evaluated in primary carcinomas and, if avail-
able, in corresponding recurrent carcinomas.
Patients were considered to have a “strong family history”of cancer
if they had a relative with OC, a relative with breast cancer before age 50,
or two relatives with breast cancer at any age.
All statistical analyses were pre-planned based on our hypothe-
ses. The Fisher's exact test was used to test the significance of
contingency tables (Tables 1 and 2). Progression-free survival (PFS)
was defined as the time between study enrollment and disease
progression or death. Patients who did not receive chemotherapy
and those for whom chemotherapy information was not available
were excluded from the PFS analysis. Overall survival (OS) was
defined as the time between study enrollment and last follow up
visit or death. Patients with stage I disease were excluded from
survival analyses. Kaplan Meier curves were generated for OS and
PFS and evaluated by the Log Rank test. The study sample size was
not large enough to correct for confounders with multivariate
analysis.
3. Results
Primary carcinomas from a total of 332 patients were evaluated.
Table 1 provides demographic information for all subjects. For 12 pa-
tients, paired recurrent neoplasms were also analyzed for methylation.
Tables 1 and 2 summarize the methylation and germline mutation
status of the primary carcinomas. Mutation information for all but
three patients was previously published [2]. Sixty-nine (20.8%) carcino-
mas had a germline mutation in one or more of the HRR genes assayed
(1 ATM,2BARD1,38BRCA1,13BRCA2,4BRIP1,3CHEK2,1NBN,1PALB2,
3RAD51C, and 4 RAD51D). Twenty-eight (8.4%) carcinomas had a so-
matic mutation in one or more HRR genes (2 ATM,16BRCA1,5BRCA2,
1BRIP1,4CHEK2,1MRE11A,1RAD51C,1SLX4). Among these mutated
cases, six (1.8%) had mutations in more than one gene. These cases
were a germline BRCA1 mutation with a somatic CHEK2 mutation, a
germline BRCA1 mutation with a somatic ATM mutation, a germline
BRCA1 mutation with a germline BARD1 mutation, a germline CHEK2
mutation with a somatic BRCA2 mutation, a somatic CHEK2 mutation
with a somatic SLX4 mutation, and a case with somatic mutations in
BRCA1,BRIP1,andMRE11A.
A total of 31 primary carcinomas were found to have methylation of
either BRCA1 (22, 6.6%) or RAD51C (9, 2.7%). Two hundred seven
(62.3%) carcinomas had neither methylation nor mutation. No carcino-
ma had both a germline mutation in a HRR gene and methylation of ei-
ther BRCA1 or RAD51C (P= 0.0008). One carcinoma with a somatic
mutation in BRIP1 had methylation of BRCA1.
When available, paired recurrent carcinomas were also evaluated for
somatic mutations and methylation. All paired carcinomas had concor-
dant methylation status: eleven of the recurrent carcinomas were
unmethylated as were their corresponding primary neoplasms, and
one recurrent carcinoma was BRCA1 methylated, as was the primary
neoplasm associated with it.
Table 1 describes the clinical features of the study population by
methylation and mutation status. At time of diagnosis, patients with
BRCA1 methylation of their carcinoma were on average 5.6 years youn-
ger (mean 57.7 years ± 2.5) than patients without germline or somatic
mutations and without methylation (mean 63.3 years ± 0.79) (P=
0.029). Patients with germline or somatic BRCA1 mutations were youn-
ger at diagnosis than patients without mutations and without methyla-
tion by 9.3 years ± 1.7 (mean 54.1 years ± 1.4) (Pb0.0001). The
difference in age at diagnosis was more pronounced when the analysis
was restricted to patients with germline BRCA1 mutations, who were on
average 10.9 years younger (mean 52.4 years ± 1.4) than patients with-
out mutations and without methylation (Pb0.0001). Patients with
germline or somatic mutations in HRR genes other than BRCA1 or
RAD51C were also younger at diagnosis than patients without muta-
tions and without methylation by 5.83 ± 2.04 years (mean 57.5 ±
1.9 years) (P= 0.005).
Germline BRCA1 mutations and BRCA1 methylation were both asso-
ciated with HGS histology when compared to carcinomas without mu-
tation or methylation (95% vs 71%, P= 0.001; 91% vs 71%, P=0.045),
but this was not true for BRCA1 somatic mutations. Of the 16 somatically
mutated BRCA1 carcinomas, eight were HGS, four were undifferentiated,
two were endometrioid, one was clear cell, and one was a carcinosarco-
ma. A strong family history of breast or ovarian carcinoma was more
common in patients with germline HRR mutations including mutations
in BRCA1 and RAD51C (55% vs 17% of patients whose carcinomas were
neither mutated nor methylated, Pb0.0001). Patients with germline
HRR gene mutations had a significantly higher incidence of a personal
history of breast cancer (21.7% vs 3.4% of those with neither mutation
nor methylation, Pb0.0001). Advanced stage disease (stage III or IV),
utilization of neoadjuvant chemotherapy, and achievement of optimal
primary surgical debulking was similar in carcinomas with or without
mutation or methylation. As previously published, germline or somatic
BRCA1 mutations were associated with increased platinum sensitivity
compared to carcinomas without mutation or methylation (80% vs
2S.S. Bernards et al. / Gynecologic Oncology xxx (2017) xxx–xxx
Please cite this article as: S.S. Bernards, et al., Clinical characteristics and outcomes of patients with BRCA1 or RAD51C methylated versus mutated
ovarian carcinoma, Gynecol Oncol (2017), https://doi.org/10.1016/j.ygyno.2017.12.004

62%, P= 0.034) [2]. Germline or somatic mutations in HRR genes other
than BRCA1 or RAD51C were also associated with increased sensitivity to
platinum chemotherapy (85% vs 62%, P= 0.015). Conversely, BRCA1
methylation was not associated with increased platinum sensitivity
(68% vs 62%, P= 0.803). Taking into account the existing platinum sen-
sitivity rate of 75% and the number of cases with missing platinum sen-
sitivity data, we had an 80% power to detect a 30% change in the fraction
of cases that are platinum sensitive comparing the methylated cases to
the non-methylated, non-mutated cases. Germline or somatic BRCA1
mutations were associated with TP53 mutations when compared to car-
cinomas without methylation or mutations (89% vs 67% P= 0.0012).
BRCA1 methylation was not associated with TP53 mutations (82% vs
67%, P= 0.23).
Fig. 1 provides survival analyses of this patient cohort divided into
carcinomas with BRCA1 or RAD51C methylation, carcinomas with
germline or somatic mutations in HRR genes, and carcinomas without
methylation or mutations. There was no significant difference in
progression-free survival between these three groups, though PFS as-
sessment was limited by missing data for many patients. Patients
whose carcinomas had HRR mutations had a statistically significant
increase in overall survival compared to those with methylated or
with unmethylated, non-mutated carcinomas. The small total number
of methylated cases (31/332, 9.3%) meant we were underpowered
to detect a difference in survival between methylated cases and
unmethylated, non-mutated cases. Median survival was 41 months in
methylated cases (HR 0.79, 95% CI 0.51–1.26), 43 months in neither mu-
tated nor methylated cases, and 66 months in mutated cases (HR 0.70,
95% CI 0.53–0.94).
4. Discussion
BRCA1 methylation was present in 6.6% (22/332) of ovarian carcino-
mas in our series. Compared to studies also unrestricted by histology,
our rate was similar to the rate reported by Cunningham et al. (9%,
Table 1
Clinicopathologic characteristics of study patients by mutation and methylation status.
Germline or somatic
BRCA1 mutation
BRCA1
methylation
Germline or somatic
RAD51C mutation
RAD51C
methylation
Germline or somatic mutations in
another HRR gene
a
No mutation or
methylation
All
patients
N 54 (16.3%) 22 (6.6%) 4 (1.2%) 9 (2.7%) 37 (11.1%)
e
207 (62.3%) 332
Median age at dx (range)
b
53 (29–76) 55 (37–85) 57.5 (47–70) 59 (39–67) 56 (34–83) 63 (34–90) 60 (29–90)
Strong family history
c
27 (50%) 7 (31.8%) 2 (50%) 3 (33.3%) 18 (50%) 36 (17.4%) 93 (28%)
Personal hx of br ca 10 (18.5%) 3 (13.6%) 0 0 6 (16.7%) 7 (3.4%) 26 (7.8%)
Neoadjuvant chemo 3 (5.6%) 2 (9.1%) 0 2 (22.2%) 4 (11.1%) 19 (9.2%) 30 (9%)
Somatic TP53 mutation 48 (88.9%) 18 (81.8%) 2 (50%) 4 (44.4%) 24 (64.9%) 138 (66.7%) 234 (70.5%)
Optimally debulked (b1 cm)
Yes 35 (64.8%) 16 (72.7%) 4 (100%) 5 (55.6%) 29 (80.6%) 141 (68.1%) 230 (69.3%)
No 18 (33.3%) 6 (27.3%) 0 4 (44.4%) 7 (19.4%) 64 (30.9%) 99 (29.8%)
Not available 1 (1.9%) 0 0 0 0 2 (1%) 3 (0.9%)
Platinum sensitivity
d
Sensitive 36 (66.7%) 13 (59.1%) 2 (50%) 3 (33.3%) 28 (77.8%) 109 (52.7%) 191 (57.5%)
Resistant 6 (11.1%) 4 (18.2%) 1 (25%) 1 (11.1%) 2 (5.6%) 31 (15%) 45 (13.5%)
Refractory 3 (5.5%) 2 (9.1%) 1 (25%) 4 (44.5%) 3 (8.3%) 35 (16.9%) 48 (14.5%)
Not available 9 (16.7%) 3 (13.6%) 0 1 (11.1%) 3 (8.3%) 32 (15.4%) 48 (14.5%)
Histology
HG serous 44 (81.5%) 20 (91%) 2 (50%) 5 (55.6%) 24 (66.7%) 146 (70.5%) 241 (72.6%)
Undifferentiated 5 (9.2%) 1 (4.5%) 1 (25%) 3 (33.3%) 4 (11.1%) 14 (6.8%) 28 (8.5%)
LG serous 0 0 0 0 1 (2.8%) 8 (3.9%) 9 (2.7%)
Endometrioid 3 (5.5%) 1 (4.5%) 0 0 3 (8.3%) 16 (7.7%) 23 (6.9%)
Clear cell 1 (1.9%) 0 0 1 (11.1%) 4 (11.1%) 10 (4.8%) 16 (4.8%)
Carcinosarcoma 1 (1.9%) 0 1 (25%) 0 0 11 (5.3%) 13 (3.9%)
Transitional 0 0 0 0 0 1 (0.5%) 1 (0.3%)
Neuroendocrine 0 0 0 0 0 1 (0.5%) 1 (0.3%)
FIGO stage
Stage I 2 (3.7%) 1 (4.5%) 0 0 5 (13.9%) 14 (6.8%) 22 (6.6%)
Stage II 1 (1.9%) 1 (4.5%) 1 (25%) 1 (11.1%) 2 (5.6%) 13 (6.3%) 19 (5.7%)
Stage III 42 (77.7%) 15 (68.2%) 1 (25%) 7 (77.8%) 26 (72.2%) 151 (72.9%) 242 (72.9%)
Stage IV 8 (14.8%) 3 (13.7%) 2 (50%) 1 (11.1%) 3 (8.3%) 29 (14%) 46 (13.9%)
Unstaged 1 (1.9%) 2 (9.1%) 0 0 0 0 3 (0.9%)
a
Mutation in: ATM,ATR,BARD1,BLM,BRCA2,BRIP1,CDK12,CHEK2,MRE11A,NBN,PALB2,RAD51D,RBBP8,SLX4,orXRCC2. All cases of somatic mutations in multiple HRR genes were
counted in the “Germline or Somatic mutations in another HRR gene”category with two exceptions. If one of the genes was BRCA1, the patient was added to the “Germline or Somatic
BRCA1 Mutation”category instead. If the patient had a somatic mutation in multiple HRR genes including RAD51C but excluding BRCA1, the patient was counted in the “Germline or So-
matic RAD51C Mutation”group.
b
There is a statistically significant difference in age at diagnosis between cases with “BRCA1 methylation”(5.6 ± 2.5 years younger) and cases with “No mutations or methylation”(P=
0.029), between cases with “Germline or Somatic BRCA1 mutation”(9.3 ± 1.7 years younger) and cases with “No mutations or methylation”(P≤0.0001), and between cases with
“Germline or somatic mutations in another HRR gene”(5.83 ± 2.04 years younger) and cases with “No mutations or methylation”(P=0.005).
c
Strong family history = a relative with OC, a relative with breast cancer b50 years of age, or two relatives with breast cancer at any age. Two “Germline or Somatic BRCA1 mutation”
patients, one “RAD51C methylation”patient, one “Germline or somatic mutations in another HRR gene”patient, and five “No mutation or methylation”patients have unknown family his-
tory due to being adopted.
d
“Germline or Somatic BRCA1 mutation”was associated with platinum sensitivity when compared to cases with “No mutation or methylation”(P= 0.034). Cases with “Germline or
somatic mutations in another HRR gene”were also associated with platinum sensitivity (P= 0.015). “BRCA1 methylation”was not associated with platinum sensitivity (P=0.803).
e
One carcinoma with a somatic BRIP1 mutation was also BRCA1 methylated and is counted in both categories in the “N”row (“BRCA1 methylated”and “Germline or somatic mutations
in another HRR gene”). It is counted only in the “BRCA1 methylated”category for the remaining rows.
Table 2
Methylation of BRCA1 or RAD51C and mutation in HRR genes in study patients.
+ for HHR mut
a
–for HRR mut
a
Total
+ for methylation 1
b
30 31
–for methylation 94 207 301
Total 95 237 332
a
HRR = Homologous Recombination Repair; mutation in: ATM,ATR,BARD1,BLM,
BRCA1,BRCA2,BRIP1,CKD12,CHEK2,MRE11A,NBN,PALB2,RAD51C,RAD51D,RBBP8,SLX4,
or XRCC2.
b
BRCA1 methylation, somatic BRIP1 mutation.
3S.S. Bernards et al. / Gynecologic Oncology xxx (2017) xxx–xxx
Please cite this article as: S.S. Bernards, et al., Clinical characteristics and outcomes of patients with BRCA1 or RAD51C methylated versus mutated
ovarian carcinoma, Gynecol Oncol (2017), https://doi.org/10.1016/j.ygyno.2017.12.004

45/482, P= 0.19) but possibly lower than Baldwin et al. (12.2%,
12/98, P=0.08)[15,16]. Two studies assessed BRCA1 methylation
only in HGS ovarian carcinomas including TCGA (11.5%, 56/489)
and Ruscito et al. (14.8%, 38/257) [1,14].Whenwerestrictedour
analyses to only our 241 HGS cases, BRCA1 methylation was
present in 20 (8.3%) carcinomas, which was similar to TCGA's
study (11.5%, P= 0.20) but lower than found by Ruscito et al.
(14.8%, P=0.03)[1,14].WedevelopedourownRAD51C methylation
sensitive PCR assay [9] after finding inconsistent results from repet-
itive experiments using the assay previously reported [25].Despite
differences in RAD51C methylation assays, we found similar rates to
that seen in TCGA (3%, P= 1) and Cunningham et al. (1%, P= 0.21)
[1,15].
Promoter hypermethylation of RAD51C and BRCA1 did not occur
in cases with damaging germline or somatic mutations in these genes
(P= 0.001). This mutual exclusivity was also evident in two large stud-
ies of methylation in ovarian carcinoma [1,15]. These data support
methylation as an alternate mechanism for HRR down regulation that
provides a similar advantage in ovarian tumorigenesis as that for HRR
mutation.
In this study, we found similarities in the clinical presentation of
BRCA1 methylated and mutated carcinomas, including younger patient
age at diagnosis and HGS histology. Ruscito et al. also found that
BRCA1 methylation was associated with younger patient age at diagno-
sis in 257 HGS ovarian carcinomas [14]. However, a larger study is need-
ed to confirm these findings. Why somatic BRCA1 methylation is
associated with younger age of diagnosis is not immediately clear, but
deserves further investigation.
Despite these similarities in clinical features, there were
important differences between BRCA1 methylated and mutated ovarian
carcinomas. BRCA1 methylation was not associated with longer overall
survival. However, our power to detect a survival difference between
BRCA1 methylated and unmethylated, non-mutated cases was limited
by the small number of BRCA1 methylated cases in this series. In con-
trast, BRCA1 mutation was associated with longer survival and sensitiv-
ity to platinum chemotherapy, as previously shown [2,5–8,24].
Similarly, TCGA's study found that patients with BRCA1 or BRCA2 mutat-
ed carcinomas had longer overall survival than those with BRCA1 meth-
ylation or without alteration of BRCA1 or BRCA2 [1]. Some studies have
shown that BRCA2 mutation carriers have improved 5-year survival
compared to BRCA1 mutation carriers [3,8]. However, these studies
were either very large [3] or studied a patient population with high
BRCA1/2 mutation rates [8]. The current study was too small to separate-
ly compare survival in BRCA1 versus BRCA2 carriers.
Similar to our findings for overall survival, we found no association
between BRCA1 and RAD51C methylation and platinum sensitivity, but
again, these analyses are limited by small numbers of methylated can-
cers. Ruscito et al. also found no significant difference in response to
platinum chemotherapy between BRCA1 methylated and unmethylated
cases, but did not account for mutation status [14]. The lack of an asso-
ciation with platinum sensitivity for cases with BRCA1 or RAD51C meth-
ylation is somewhat surprising as methylation of these genes in
recurrent ovarian cancer has recently been associated with a high re-
sponse to the PARP inhibitor rucaparib, suggesting that methylation
confers a homologous recombination deficient phenotype [9].BRCA1
and RAD51C promoter hypermethylation have also been recently asso-
ciated with high genomic loss of heterozygosity (LOH), consistent
with homologous recombination deficiency [9].Itispossiblethat
some cases have only hemi-allelic BRCA1 promoter methylation with-
out associated loss of heterozygosity, which would not be expected to
impact therapeutic sensitivity. The lack of a survival advantage in pa-
tients with methylated ovarian carcinomas could also result from rela-
tively rapid loss of methylation during treatment with platinum
chemotherapy, as loss of methylation has been recently described in
platinum and PARP inhibitor resistant ovarian carcinoma [25]. Alterna-
tively, treatment with platinum chemotherapy could select for
unmethylated carcinoma cells among an initially heterogeneous popu-
lation. Unfortunately, we had few paired cases to test these hypotheses,
and the only methylated primary carcinoma with a paired recurrent
sample was also methylated in the recurrence. For that case, both the
methylated primary carcinoma and the methylated paired recurrence
responded well to platinum therapy. Baldwin et al. evaluated matched
recurrent samples for two BRCA1 methylated primary cases, both of
which were also methylated in the recurrence [16]. Patch et al. found
one case in which the primary, platinum sensitive carcinoma was
BRCA1 methylated while the platinum resistant recurrence was
unmethylated [25]. In a larger series of 13 BRCA1 methylated primary
ovarian carcinomas, 4 (31%) were unmethylated in a paired recurrent
biopsy, though all were predicted to be platinum sensitive based on in-
terval to recurrence [9].
Technical advances and declining costs have made sequencing
patients' carcinomas more feasible. Genetic and genomic markers
to direct anti-neoplastic therapies are becoming increasingly com-
mon; the poly ADP-ribose polymerase (PARP) inhibitor rucaparib
was recently approved for treatment of recurrent ovarian cancer
with germline and somatic mutations, making BRCA1 and BRCA2 mu-
tations the first “actionable”somatic mutations in ovarian cancer as-
sociated with an approved, targeted therapy [26]. The role of
promoter methylation (or loss of methylation) of specificgenes
and relationship to sensitivity or resistance to various therapies de-
serves additional study. Our data suggest that BRCA1 and RAD51C
methylation are not associated with improved outcomes or with
platinum sensitivity in patients with unselected primary ovarian
carcinomas.
Mutation Category Hazard Ratio (95% CI) P-value
RAD51C and BRCA1 Methylation 0.79 (0.51-1.26) 0.3
Mutation in any HR gene 0.70 (0.53-0.94) 0.02
Percent survival
Mutation Category Hazard Ratio (95% CI) P-value
RAD51C and BRCA1 Methylation 0.80 (0.51-1.27) 0.36
Mutation in any HR gene 0.76 (0.56-1.02) 0.07
0 50 100 150 200 250
0
50
100
Months
Percent survival
RAD51C and BRCA1 methylation (18)
Mutation in any HR gene (25)
No mutation, no methylation (17)
Median Months PFS
Fig. 1. Overall and progression-free survival by mutation and methylation status.
4S.S. Bernards et al. / Gynecologic Oncology xxx (2017) xxx–xxx
Please cite this article as: S.S. Bernards, et al., Clinical characteristics and outcomes of patients with BRCA1 or RAD51C methylated versus mutated
ovarian carcinoma, Gynecol Oncol (2017), https://doi.org/10.1016/j.ygyno.2017.12.004

Conflict of interest statement
The authors have no conflicts of interest to declare.
Funding
University of Washington School of Medicine Medical Student Research Training Program
(MSRTP) [SSB], Stand Up To Cancer –Ovarian Cancer Research Fund Alliance –National
Ovarian Cancer Coalition Dream Team Translational Research Grant (Grant Number:
SU2C-AACR-DT16-15) [EMS]. Stand Up to Cancer is a program of the Entertainment Indus-
try Foundation; research grants are administered by the American Association for Cancer
Research, a scientific partner of Stand Up To Cancer. V Foundation Translational Research
Award [EMS].
References
[1] D. Bell, et al., Integrated genomic analyses of ovarian carcinoma, Nature 474 (7353)
(2011) 609–615.
[2] K.P. Pennington, et al., Germline and somatic mutations in homologous recombina-
tion genes predict platinum response and survival in ovarian, fallopian tube, and
peritoneal carcinomas, Clin. Cancer Res. 20 (3) (2014) 764–775.
[3] B.M. Norquist, et al., Inherited mutations in women with ovarian carcinoma, JAMA
Oncol. 2 (4) (2016) 482–490.
[4] S. Zhang, et al., Frequencies of BRCA1 and BRCA2 mutations among 1,342 unselected
patients with invasive ovarian cancer, Gynecol. Oncol. 121 (2) (2011) 353–357.
[5] P.M.L.H. Vencken, et al., Chemosensitivity and outcome of BRCA1- and BRCA2-
associated ovarian cancer patients after first-line chemotherapy compared with
sporadic ovarian cancer patients, Ann. Oncol. 22 (6) (2011) 1346–1352.
[6] D. Yang, et al., Association of BRCA1 and BRCA2 mutations with survival, chemo-
therapy sensitivity, and gene mutator phenotype in patients with ovarian cancer,
JAMA 306 (14) (2011) 1557–1565.
[7] K.L. Bolton, et al., Association between BRCA1 and BRCA2 mutations and survival in
women with invasive epithelial ovarian cancer, JAMA 307 (4) (2012) 382–390.
[8] A. Chetrit, et al., Effect of BRCA1/2 mutations on long-term survival of patients with
invasive ovarian cancer: the National Israeli Study of ovarian cancer, J. Clin. Oncol.
26 (1) (2008) 20–25.
[9] E.M. Swisher, et al., Rucaparib in relapsed, platinum-sensitive high-grade ovarian
carcinoma (ARIEL2 part 1): an international, multicentre, open-label, phase 2 trial,
Lancet Oncol. 18 (1) (2017) 75–87.
[10] J.A. Ledermann, et al., Overall survival in patients with platinum-sensitive recurrent
serous ovarian cancer receiving olaparib maintenance monotherapy: an updated
analysis from a randomised, placebo-controlled, double-blind, phase 2 trial, Lancet
Oncol. 17 (11) (2016) 1579–1589.
[11] M.R. Mirza, et al., Niraparib maintenance therapy in platinum-sensitive, recurrent
ovarian cancer, N. Engl. J. Med. 375 (22) (2016) 2154–2164.
[12] P.A. Jones, S.B. Baylin, The fundamental role of epigenetic events in cancer, Nat. Rev.
Genet. 3 (6) (2002) 415–428.
[13] J.P. Geisler, et al., Frequency of BRCA1 dysfunction in ovarian cancer, J. Natl. Cancer
Inst. 94 (1) (2002) 61–67.
[14] I. Ruscito, et al., BRCA1 gene promoter methylation status in high-grade serous ovar-
ian cancer patients–a study of the tumour Bank ovarian cancer (TOC) and ovarian
cancer diagnosis consortium (OVCAD), Eur. J. Cancer 50 (12) (2014) 2090–2098.
[15] J.M. Cunningham, et al., Clinical characteristics of ovarian cancer classified by BRCA1,
BRCA2, and RAD51C status, Sci. Rep. 4 (2014) 4026.
[16] R.L. Baldwin, et al., BRCA1 promoter region hypermethylation in ovarian carcinoma:
a population-based study, Cancer Res. 60 (19) (2000) 5329–5333.
[17] M. Esteller, et al., Promoter hypermethylation and BRCA1 inactivation in sporadic
breast and ovarian tumors, J. Natl. Cancer Inst. 92 (7) (2000) 564–569.
[18] C. Wang, et al., Expression of BRCA1 protein in benign, borderline, and malignant
epithelial ovarian neoplasms and its relationship to methylation and allelic loss of
the BRCA1 gene, J. Pathol. 202 (2) (2004) 215–223.
[19] E.M. Swisher, et al., Methylation and protein expression of DNA repair genes: asso-
ciation with chemotherapy exposure and survival in sporadic ovarian and peritone-
al carcinomas, Mol. Cancer 8 (2009) 48.
[20] J.W. Chiang, et al., BRCA1 promoter methylation predicts adverse ovarian cancer
prognosis, Gynecol. Oncol. 101 (3) (2006) 403–410.
[21] O.A. Stefansson, et al., BRCA1 epigenetic inactivation predicts sensitivity to
platinum-based chemotherapy in breast and ovarian cancer, Epigenetics 7 (11)
(2012) 1225–1229.
[22] T. Walsh, et al., Detection of inherited mutations for breast and ovarian cancer using
genomic capture and massively parallel sequencing, Proc. Natl. Acad. Sci. U. S. A. 107
(28) (2010) 12629–12633.
[23] S.S. Bernards, et al., Genetic characterization of early onset ovarian carcinoma,
Gynecol. Oncol. 140 (2) (2016) 221–225.
[24] K. Alsop, C. Meldrum, A. de Fazio, C. Emmanuel, J. George, et al., BRCA mutation fre-
quency and patterns of treatment response in BRCA mutation-positive women with
ovarian cancer: a report from the Australian Ovarian Cancer Study Group, J. Clin.
Oncol. 30 (21) (2012) 2654–2663.
[25] A.-M. Patch, et al., Whole–genome characterization of chemoresistant ovarian can-
cer, Nature 521 (7553) (2015) 489–494.
[26] Databases. U.F.a.D.A.D.A.a. Rucaparib, Available from: https://www.fda.gov/Drugs/
InformationOnDrugs/ApprovedDrugs/ucm533891.htm 2016, Accessed date: 19 De-
cember 2016.
5S.S. Bernards et al. / Gynecologic Oncology xxx (2017) xxx–xxx
Please cite this article as: S.S. Bernards, et al., Clinical characteristics and outcomes of patients with BRCA1 or RAD51C methylated versus mutated
ovarian carcinoma, Gynecol Oncol (2017), https://doi.org/10.1016/j.ygyno.2017.12.004
1
/
5
100%
![Poster LIBER san antonio 2011 [Mode de compatibilité]](http://s1.studylibfr.com/store/data/000441925_1-0f624c1012097e18f69fca01a2951eb6-300x300.png)